We’ve studied the Fourier transform quite a bit on this blog: with four primers and the Fast Fourier Transform algorithm under our belt, it’s about time we opened up our eyes to higher dimensions.
Indeed, in the decades since Cooley & Tukey’s landmark paper, the most interesting applications of the discrete Fourier transform have occurred in dimensions greater than 1. But for all our work we haven’t yet discussed what it means to take an “n-dimensional” Fourier transform. Our past toiling and troubling will pay off, though, because the higher Fourier transform and its 1-dimensional cousin are quite similar. Indeed, the shortest way to describe the $ n$-dimensional transform is as the 1-dimensional transform with inner products of vector variables replacing regular products of variables.
In this post we’ll flush out these details. We’ll define the multivariable Fourier transform and it’s discrete partner, implement an algorithm to compute it (FFT-style), and then apply the transform to the problem of digitally watermarking images.
As usual, all the code, images, and examples used in this post are available on this blog’s Github page.
Sweeping Some Details Under the Rug
We spent our first and second primers on Fourier analysis describing the Fourier series in one variable, and taking a limit of the period to get the Fourier transform in one variable. By all accounts, it was a downright mess of notation and symbol manipulation that culminated in the realization that the Fourier series looks a lot like a Riemann sum. So it was in one dimension, it is in arbitrary dimension, but to save our stamina for the applications we’re going to treat the $ n$-dimensional transform differently. We’ll use the 1-dimensional transform as a model, and magically generalize it to operate on a vector-valued variable. Then the reader will take it on faith that we could achieve the same end as a limit of some kind of multidimensional Fourier series (and all that nonsense with Schwarz functions and tempered distributions is left to the analysts), or if not we’ll provide external notes with the full details.
So we start with a real-valued (or complex-valued) function $ f : \mathbb{R}^n \to \mathbb{R}$, and we write the variable as $ x = (x_1, \dots, x_n)$, so that we can stick to using the notation $ f(x)$. Rather than think of the components of $ x$ as “time variables” as we did in the one-dimensional case, we’ll usually think of $ x$ as representing physical space. And so the periodic behavior of the function $ f$ represents periodicity in space. On the other hand our transformed variables will be “frequency” in space, and this will correspond to a vector variable $ \xi = (\xi_1, \dots, \xi_n)$. We’ll come back to what the heck “periodicity in space” means momentarily.
Remember that in one dimension the Fourier transform was defined by
$ \displaystyle \mathscr{F}f(s) = \int_{-\infty}^\infty e^{-2\pi ist}f(t) dt$.
And it’s inverse transform was
$ \displaystyle \mathscr{F}^{-1}g(t) = \int_{-\infty}^\infty e^{2\pi ist}f(s) ds$.
Indeed, with the vector $ x$ replacing $ t$ and $ \xi$ replacing $ s$, we have to figure out how to make an analogous definition. The obvious thing to do is to take the place where $ st$ is multiplied and replace it with the inner product of $ x$ and $ \xi$, which for this post I’ll write $ x \cdot \xi$ (usually I write $ \left \langle x, \xi \right \rangle$). This gives us the $ n$-dimensional transform
$ \displaystyle \mathscr{F}f(\xi) = \int_{\mathbb{R}^n} e^{-2\pi i x \cdot \xi}f(x) dx$,
and its inverse
$$\displaystyle \mathscr{F}^{-1}g(t) = \int_{\mathbb{R}^n} e^{2\pi i x \cdot \xi}f( \xi ) d \xi$$Note that the integral is over all of $ \mathbb{R}^n$. To give a clarifying example, if we are in two dimensions we can write everything out in coordinates: $ x = (x_1, x_2), \xi = (\xi_1, \xi_2)$, and the formula for the transform becomes
$ \displaystyle \mathscr{F}f(\xi_1, \xi_2) = \int_{-\infty}^{\infty} \int_{-\infty}^{\infty} e^{-2 \pi i (x_1 \xi_1 + x_2 \xi_2)} f(\xi_1, \xi_2) dx_1 dx_2$.
Now that’s a nasty integral if I’ve ever seen one. But for our purposes in this post, this will be as nasty as it gets, for we’re primarily concerned with image analysis. So representing things as vectors of arbitrary dimension is more compact, and we don’t lose anything for it.
Periodicity in Space? It’s All Mostly the Same
Because arithmetic with vectors and arithmetic with numbers is so similar, it turns out that most of the properties of the 1-dimensional Fourier transform hold in arbitrary dimension. For example, the duality of the Fourier transform and its inverse holds, because for vectors $ e^{-2 \pi i x \cdot (-\xi)} = e^{2 \pi i x \cdot \xi}$. So just like in on dimension, we have
$$\mathscr{F}f(-\xi) = \mathscr{F}^{-1}f(\xi)$$And again we have correspondences between algebraic operations: convolution in the spatial domain corresponds to convolution in the frequency domain, the spectrum is symmetric about the origin, etc.
At a more geometric level, though, the Fourier transform does the same sort of thing as it did in the one-dimensional case. Again the complex exponentials form the building blocks of any function we want, and performing a Fourier transform on an $ n$-dimensional function decomposes that function into its frequency components. So a function that is perfectly periodic corresponds to a Fourier spectrum that’s perfectly concentrated at a point.
But what the hell, the reader might ask, is ‘periodicity in space’? Since we’re talking about images anyway, the variables we care about (the coordinates of a pixel) are spatial variables. You could, if you were so inclined, have a function of multiple time variables, and to mathematicians a physical interpretation of dimension is just that, an interpretation. But as confusing as it might sound, it’s actually not so hard to understand the Fourier transform when it’s specialized to image analysis. The idea is that complex exponentials $ e^{\pm 2 \pi i s \cdot \xi}$ oscillate in the $ x$ variable for a fixed $ \xi$ (and since $ \mathscr{F}$ has $ \xi$ as its input, we do want to fix $ \xi$). The brief mathematical analysis goes like this: if we fix $ \xi$ then the complex exponential is periodic with magnitudinal peaks along parallel lines spaced out at a distance of $ 1/ \left \| \xi \right \|$ apart. In particular any image is a sum of a bunch of these “complex exponential with a fixed $ \xi$” images that look like stripes with varying widths and orientations (what you see here is just the real part of a particular complex exponential).
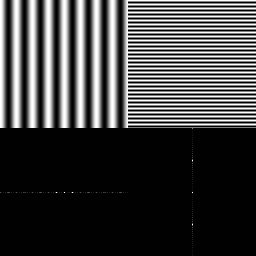
Any image can be made from a sum of a whole lot of images like this one. This corresponds to a single point in the Fourier spectrum.
What you see on top is an image, and on bottom its Fourier spectrum. That is, each brightly colored pixel corresponds to a point $ [x_1, x_2]$ with a large magnitude for that frequency component $ |\mathscr{F}f[x_1, x_2]|$.
It might be a bit surprising that every image can be constructed as a sum of stripey things, but so was it that any sound can be constructed as a sum of sines and cosines. It’s really just a statement about a basis of some vector space of functions. The long version of this story is laid out beautifully in pages 4 – 7 of these notes. The whole set of notes is wonderful, but this section is mathematically tidy and needs no background; the remainder of the notes outline the details about multidimensional Fourier series mentioned earlier, as well as a lot of other things. In higher dimensions the “parallel lines” idea is much the same, but with lines replaced by hyperplanes normal to the given vector.
Discretizing the Transform
Recall that for a continuous function $ f$ of one variable, we spent a bit of time figuring out how to find a good discrete approximation of $ f$, how to find a good discrete approximation of the Fourier transform $ \mathscr{F}f$, and how to find a quick way to transition between the two. In brief: $ f$ was approximated by a vector of samples $ (f[0], f[1], \dots, f[N])$, reconstructed the original function (which was only correct at the sampled points) and computed the Fourier transform of that, calling it the discrete Fourier transform, or DFT. We got to this definition, using square brackets to denote list indexing (or vector indexing, whatever):
Definition: Let $ f = (f[1], \dots f[N])$ be a vector in $ \mathbb{R}^N$. Then the discrete Fourier transform of $ f$ is defined by the vector $ (\mathscr{F}f[1], \dots, \mathscr{F}f[N])$, where
$$\displaystyle \mathscr{F}f[j] = \sum_{k=0}^{N-1} f[k]e^{-2 \pi i jk/N}$$Just as with the one-dimensional case, we can do the same analysis and arrive at a discrete approximation of an $ n$-dimensional function. Instead of a vector it would be an $ N \times N \times \dots \times N$ matrix, where there are $ n$ terms in the matrix, one for each variable. In two dimensions, this means the discrete approximation of a function is a matrix of samples taken at evenly-spaced intervals in both directions.
Sticking with two dimensions, the Fourier transform is then a linear operator taking matrices to matrices (which is called a tensor if you want to scare people). It has its own representation like the one above, where each term is a double sum. In terms of image analysis, we can imagine that each term in the sum requires us to look at every pixel of the original image
Definition: Let $ f = (f[s,t])$ be a vector in $ \mathbb{R}^N \times \mathbb{R}^M$, where $ s$ ranges from $ 0, \dots, N-1$ and $ t$ from $ 0, \dots, M-1$. Then the discrete Fourier transform of $ f$ is defined by the vector $ (\mathscr{F}f[s,t])$, where each entry is given by
$$\displaystyle \mathscr{F}f[x_1, x_2] = \sum_{s=0}^{N-1} \sum_{t=0}^{M-1} f[s, t] e^{-2 \pi i (s x_1 / N + t x_2 / M)}$$In the one-dimensional case the inverse transform had a sign change in the exponent and an extra $ 1/N$ normalization factor. Similarly, in two dimensions the inverse transform has a normalization factor of $ 1/NM$ (1 over the total number of samples). Again we use a capital $ F$ to denote the transformed version of $ f$. The higher dimensional transforms are analogous: you get $ n$ sums, one for each component, and the normalization factor is the inverse of the total number of samples.
$$\displaystyle \mathscr{F}^{-1}F[x_1, x_2] = \frac{1}{NM} \sum_{s=0}^{N-1} \sum_{t=0}^{M-1} f[s,t] e^{2 \pi i (sx_1 / N + tx_2 / M)}$$Unfortunately, the world of the DFT disagrees a lot on the choice of normalization factor. It turns out that all that really matters is that the exponent is negated in the inverse, and that the product of the constant terms on both the transform and its inverse is $ 1/NM$. So some people will normalize both the Fourier transform and its inverse by $ 1/ \sqrt{NM}$. The reason for this is that it makes the transform and its inverse more similar-looking (it’s just that, cosmetic). The choice of normalization isn’t particularly important for us, but beware: non-canonical choices are out there, and they do affect formulas by adding multiplicative constants.
The Fast Fourier Transform, Revisited
Now one might expect that there is another clever algorithm to drastically reduce the runtime of the 2-dimensional DFT, akin to the fast Fourier transform algorithm (FFT). But actually there is almost no additional insight required to understand the “fast” higher dimensional Fourier transform algorithm, because all the work was done for us in the one dimensional case.
All that we do is realize that each of the inner summations is a 1-dimensional DFT. That is, if we write the inner-most sum as a function of two parameters
$$\displaystyle g(s, x_2) = \sum_{t=0}^{M-1} f(s,t) e^{-2 \pi i (tx_2 / M)}$$then the 2-dimensional FFT is simply
$$\displaystyle \mathscr{F}f[x_1, x_2] = \sum_{s=0}^{N-1} g(s, x_2) e^{-2 \pi i (sx_1/N)}$$But now notice, that we can forget that $ g(s,x_2)$ was ever a separate, two-dimensional function. Indeed, since it only depends on the $ x_2$ parameter from out of the sum this is precisely the formula for a 1-dimensional DFT! And so if we want to compute the 2-dimensional DFT using the 1-dimensional FFT algorithm, we can compute the matrix of 1-dimensional DFT entries for all choices of $ s, x_2$ by fixing each value of $ s$ in turn and running FFT on the resulting “column” of values. If you followed the program from our last FFT post, then the only difficulty is in understanding how the data is shuffled around and which variables are fixed during the computation of the sub-DFT’s.
To remedy the confusion, we give an example. Say we have the following 3×3 matrix whose DFT we want to compute. Remember, these values are the sampled values of a 2-variable function.
$$\displaystyle \begin{pmatrix} f[0,0] & f[0,1] & f[0,2] \\\ f[1,0] & f[1,1] & f[1,2] \\\ f[2,0] & f[2,1] & f[2,2] \end{pmatrix}$$The first step in the algorithm is to fix a choice of row, $ s$, and compute the DFT of the resulting row. So let’s fix $ s = 0$, and then we have the resulting row
$$\displaystyle f_0 = (f[0,0], f[0,1], f[0,2])$$It’s DFT is computed (intentionally using the same notation as the inner summation above), as
$$\displaystyle g[0,x_2] = (\mathscr{F}f_0)[x_2] = \sum_{t=0}^{M-1} f_0[t] e^{- 2 \pi i (t x_2 / M)}$$Note that $ f_0[t] = f[s,t]$ for our fixed choice of $ s=0$. And so if we do this for all $ N$ rows (all 3 rows, in this example), we’ll have performed $ N$ FFT’s of size $ M$ to get a matrix of values
$$\displaystyle \begin{pmatrix} g[0,0] & g[0,1] & g[0,2] \\\ g[1,0] & g[1,1] & g[1,2] \\\ g[2,0] & g[2,1] & g[2,2] \end{pmatrix}$$Now we want to compute the rest of the 2-dimensional DFT to the end, and it’s easy: now each column consists of the terms in the outermost sum above (since $ s$ is the iterating variable). So if we fix a value of $ x_2$, say $ x_2 = 1$, we get the resulting column
$$\displaystyle g_1 = (g[0, 1], g[1,1], g[2,1])$$and computing a DFT on this row gives
$ \displaystyle \mathscr{F}f[x_1, 1] = \sum_{s=0}^{N-1} g_1[s] e^{-2 \pi i sx_1 / N}$.
Expanding the definition of $ g$ as a DFT gets us back to the original formula for the 2-dimensional DFT, so we know we did it right. In the end we get a matrix of the computed DFT values for all $ x_1, x_2$.
Let’s analyze the runtime of this algorithm: in the first round of DFT’s we computed $ N$ DFT’s of size $ M$, requiring a total of $ O(N M \log M)$, since we know FFT takes time $ O(M \log M)$ for a list of length $ M$. In the second round we did it the other way around, computing $ M$ DFT’s of size $ N$ each, giving a total of
$$O(NM \log M + NM \log N) = O(NM (\log N + \log M)) = O(NM \log (NM))$$In other words, if the size of the image is $ n = NM$, then we are achieving an $ O(n \log n)$-time algorithm, which was precisely the speedup that the FFT algorithm gave us for one-dimension. We also know a lower bound on this problem: we can’t do better than $ NM$ since we have to look at every pixel at least once. So we know that we’re only a logarithmic factor away from a trivial lower bound. And indeed, all other known DFT algorithms have the same runtime. Without any assumptions on the input data (or any parallelization), nobody knows of a faster algorithm.
Now let’s turn to the code. If we use our FFT algorithm from last time, the pure Python one (read: very slow), then we can implement the 2D Fourier transform in just two lines of Python code. Full disclosure: we left out some numpy stuff in this code for readability. You can view the entire source file on this blog’s Github page.
def fft2d(matrix):
fftRows = [fft(row) for row in matrix]
return transpose([fft(row) for row in transpose(fftRows)])
And we can test it on a simple matrix with one nonzero value in it:
A = [[0,0,0,0], [0,1,0,0], [0,0,0,0], [0,0,0,0]]
for row in fft2d(A):
print(', '.join(['%.3f + %.3fi' % (x.real, x.imag) for x in row]))
The output is (reformatted in LaTeX, obviously):
$$\displaystyle \begin{pmatrix} 1 & -i & -1 & i \\\ -i & -1 & i & 1 \\\ -1 & i & 1 & -i \\\ i & 1 & -i & -1 \end{pmatrix}$$The reader can verify by hand that this is correct (there’s only one nonzero term in the double sum, so it just boils down to figuring out the complex exponential $ e^{2 \pi i (x_1 + x_2 / 4)}$). We leave it as an additional exercise to the reader to implement the inverse transform, as well as to generalize this algorithm to higher dimensional DFTs.
Some Experiments and Animations
As we did with the 1-dimensional FFT, we’re now going to switch to using an industry-strength FFT algorithm for the applications. We’ll be using the numpy library and its “fft2” function, along with scipy’s ndimage module for image manipulation. Getting all of this set up was a nightmare (thank goodness for people who guide users like me through this stuff, but even then the headache seemed unending!). As usual, all of the code and images used in the making of this post is available on this blog’s Github page.
And so we can start playing with a sample image, a still from one of my favorite television shows:

sherlock
The Fourier transform of this image (after we convert it to grayscale) can be computed in python:
def fourierSpectrumExample(filename):
A = ndimage.imread(filename, flatten=True)
unshiftedfft = numpy.fft.fft2(A)
spectrum = numpy.log10(numpy.absolute(unshiftedfft) + numpy.ones(A.shape))
misc.imsave("%s-spectrum-unshifted.png" % (filename.split('.')[0]), spectrum)
With the result:

sherlock-spectrum-unshifted
A few notes: we use the ndimage library to load the image and flatten the colors to grayscale. Then, after we compute the spectrum, we shift and take a logarithm. This is because the raw spectrum values are too massive; plotting them without modification makes the image contrast too high.
Something is odd, though, because the brightest regions are on the edges of the image, where we might expect the highest-frequency elements to be. Actually, it turns out that a raw DFT (as computed by numpy, anyhow) is “shifted.” That is, the indices are much like they were in our original FFT post, so that the “center” of the spectrum (the lowest frequency component) is actually in the corner of the image array.
The numpy folks have a special function designed to alleviate this called fftshift. Applying it before we plot the image gives the following spectrum:

sherlock-spectrum
Now that’s more like it. For more details on what’s going on with shifting and how to use the shifting functions, see this matlab thread. (As a side note, the “smudges” in this image are interesting. We wonder what property of the original image contributes to the smudges)
Shifted or unshifted, this image represents the frequency spectrum of the image. In other words, we could take the inverse DFT of each pixel (and its symmetric partner) of this image separately, add them all together, and get back to our original image! We did just that using a different image (one of size 266 x 189, requiring a mere 25137 frequency components), to produce this video of the process:
Many thanks to James Hance for his relentlessly cheerful art.
Digital Watermarking
Now we turn to the main application of Fourier transforms to this post, the task of adding an invisible digital watermark to an image. Just in case the reader lives in a cave, a watermark is a security device used to protect the ownership or authenticity of a particular good. Usually they’re used on money to prevent counterfeits, but they’re often applied to high-resolution images on the web to protect copyrights. But perhaps more than just protect existing copyrights, watermarks as they’re used today are ugly, and mostly prevent people from taking the image (paid for or not) in the first place. Here’s an example from a big proponent of ugly watermarks, Shutterstock.com.

stock-photo
Now if you were the business of copyright litigation, you’d make a lot of money by suing people who took your clients’ images without permission. So rather than prevent people from stealing in the first place, you could put in an invisible watermark into all of your images and then crawl the web looking for stolen images with your watermark. It would be easy enough to automate (Google already did most of the work for you, if you just want to use Google’s search by image feature).
Now I’m more on the side of Fair Use For All, so I wouldn’t hope for a company to actually implement this and make using the internet that much scarier of a place. But the idea makes for an interesting thought experiment and blog post. The idea is simply to modify the spectrum of an image by adding in small, artificial frequency components. That is, the watermarked image will look identical to the original image to a human, but the Fourier spectrum will contain suspicious entries that we can extract if we know where to look.
Implementing the watermarking feature is quite easy, so let’s do that first. Let’s work again with James Hance’s fine artwork.
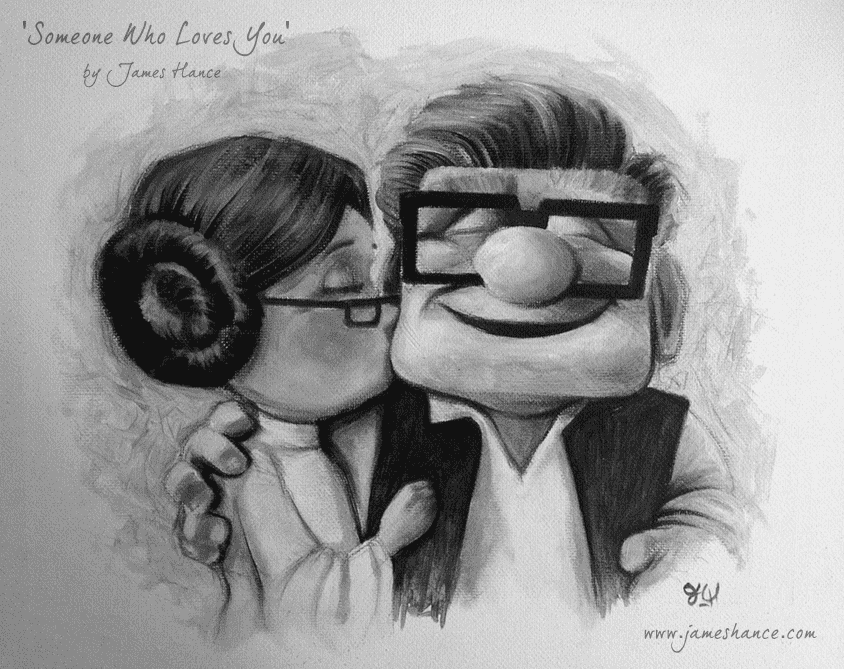
hance-up-sw-bw
Let’s call our image’s pixel matrix $ A$ and say we’re working with grayscale images for simplicity (for color, we just do the same thing to all three color channels). Then we can define a watermark matrix $ W$ by the following procedure:
- Pick a radius $ r$, a length $ L$, a watermark strength $ \alpha$, and a secret key $ k$.
- Using $ k$ as a seed to a random number generator, define a random binary vector $ v$ of length $ L$.
- Pick a subset $ S$ of the circle of coordinates centered at the image’s center of radius $ r$, chosen or rejected based on the entries of $ v$.
- Let $ W$ be the matrix of all zeros (of the same dimension as $ A$ with 1’s in the entries of $ S$.
- Compute the watermarked image as $ \mathscr{F}^{-1}(\mathscr{F}(A) + \alpha W)$. That is, compute the DFT of $ A$, add $ \alpha W$ to it, and then compute the inverse Fourier transform of the result.
The code for this is simple enough. To create a random vector:
import random
def randomVector(seed, length):
random.seed(secretKey)
return [random.choice([0,1]) for _ in range(length)]
To make the watermark (and flush out all of the technical details of how it’s done:
def makeWatermark(imageShape, radius, secretKey, vectorLength=50):
watermark = numpy.zeros(imageShape)
center = (int(imageShape[0] / 2) + 1, int(imageShape[1] / 2) + 1)
vector = randomVector(secretKey, vectorLength)
x = lambda t: center[0] + int(radius * math.cos(t * 2 * math.pi / vectorLength))
y = lambda t: center[1] + int(radius * math.sin(t * 2 * math.pi / vectorLength))
indices = [(x(t), y(t)) for t in range(vectorLength)]
for i,location in enumerate(indices):
watermark[location] = vector[i]
return watermark
We use the usual parameterization of the circle as $ t \mapsto (\cos(2 \pi t / n), \sin(2 \pi t / n)$ scaled to the appropriate radius. Here’s what the watermark looks like as a spectrum:

watermark-spectrum
And then applying a given watermark to an image is super simple.
def applyWatermark(imageMatrix, watermarkMatrix, alpha):
shiftedDFT = fftshift(fft2(imageMatrix))
watermarkedDFT = shiftedDFT + alpha * watermarkMatrix
watermarkedImage = ifft2(ifftshift(watermarkedDFT))
return watermarkedImage
And that’s all there is to it! One might wonder how the choice of $ \alpha$ affects the intensity of the watermark, and indeed here we show a few example values of this method applied to Hance’s piece:
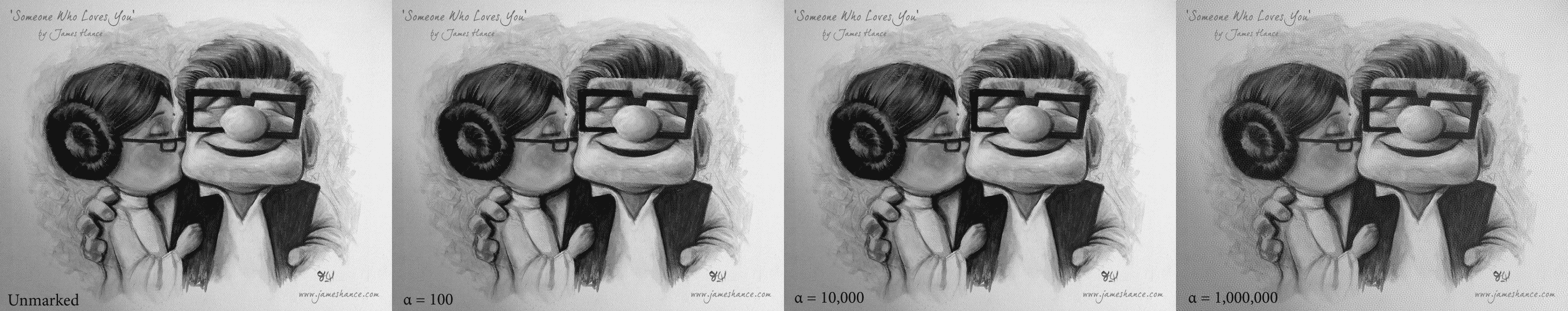
Click to enlarge. It appears that it's not until alpha becomes egregiously large that we notice the effects. This could be in part due to the fact that this is an image of a canvas (which has lots of small textures in the background).
It appears that it’s not until $ \alpha$ becomes egregiously large (over 10,000) that we visibly notice the effects. This could be in part due to the fact that this is an image of a canvas (which has lots of small textures in the background). But it’s good to keep in mind the range of acceptable values when designing a decoding mechanism.
Indeed, a decoding mechanism is conceptually much messier; it’s the art to the encoding mechanism’s science. This paper details one possible way to do it, which is essentially to scale everything up or down to 512×512 pixels and try circles of every possible radius until you find one (or don’t) which is statistically similar to the your random vector. And note that since we have the secret key we can generate the exact same random vector. So what the author of that paper suggests is to extract each circle of pixels from the Fourier spectrum, treating it as a single vector with first entry at angle 0. Then you do some statistical magic (compute cross-correlation or some other similarity measure) between the extracted pixels and your secret-key-generated random vector. If they’re sufficiently similar, then you’ve found your watermark, and otherwise there’s no watermark present.
The code required to do this only requires a few extra lines that aren’t present in the code we’re already presented in this article (numpy does cross-correlation for you), so we leave it as an exercise to the reader: write a program that determines if an image contains our watermark, and test the algorithm on various $ \alpha$ and with modifications of the image like rotation, scaling, cropping, and jpeg compression. Part of the benefit of Fourier-based techniques is the resilience of the spectrum to mild applications of these transformations.
Next time we’ll use the Fourier transform to do other cool things to images, like designing filters and combining images in interesting ways.
Until then!
Want to respond? Send me an email, post a webmention, or find me elsewhere on the internet.